Functional analysis of SdsA1, an enzyme capable of degrading SDS
Students: Avery Zierk, James Holt, Calynn Johnson, Nathan Adamson, William Zierenberg
Elucidating the mechanisms by which enzymes - macromolecules that accelerate chemical reactions by preferentially stabilizing transition states relative to ground states- control chemical reactivity is among the most exciting challenges in contemporary life sciences. To comprehend cells' inner workings it is particularly important to understand how enzymes fine tune their ability to bind specifically their reaction partners and accelerate only the desired reaction. Models derived from crystals of enzymes represent a formidable tool to visualize the three-dimensional arrangement of functional groups and the constellation of residues and putative catalytic interactions that surround the substrate molecule(s). However, models derived from crystal structures are not necessarily representative of the conformation adopted by the enzyme at the transition state of the reaction and cannot establish the energetic contributions of the different catalytic interactions.
In this project, we will investigate the mechanism of the reaction catalyzed by the alkylsulfatase SdsA1, used by the pathogen Pseudomonas aeruginosa to survive in detergents, by testing and extending the published structural model of this enzyme [1] by using site-directed mutagenesis, isotope-labeled compounds, and alternative substrates. These experiments will provide the foundation for subsequent experiments aimed to decipher the energetics involved in individual interactions, the role of specific residues in catalysis, and the specificity of SdsA1 towards its natural substrate. Because SdsA1 belongs to the zinc-metallo-hydrolases of the metallo-?-lactamase superfamily, a class of proteins comprised of enzymes with similar active site but with diverse reaction specificities [2], work done on this enzyme will provide a starting point for a work aimed to determine the factors involved in these diverse reaction specificities within the superfamily. Further, understanding of SdsA1 catalysis may have implications for the engineering of enzymes involved in bioremediation and for the development of inhibitors to fight Pseudomonas ability to survive on detergents.
[1] Hagelueken, G.; Adams, T. M.; Wiehlmann, L.; Widow, U.; Kolmar, H.; Tummler, B.; Heinz, D. W.; Schubert, W.-D. Proc Natl Acad Sci U S A 2006, 103, 7631.
[2] Daiyasu, H.; Osaka, K.; Ishino, Y.; Toh, H. FEBS Lett 2001, 503, 1
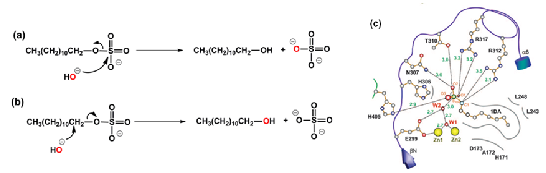
Two possible mechanisms of the reaction: S-attack (a) and C-attack (b). The two mechanisms give the same products but the use of water with a labeled oxygen atom (represented in red) allows discrimination between the two. (c) Schematic representation of the active site of SdsA1 with a bound inhibitor. Hydrogen bonds and salt bridges are marked by dotted lines, hydrophobic interactions are marked by gray arcs, and distances are in angstroms. DA is 1-decane sulfonic acid. Figure (c) is from reference 1.